Research Overview
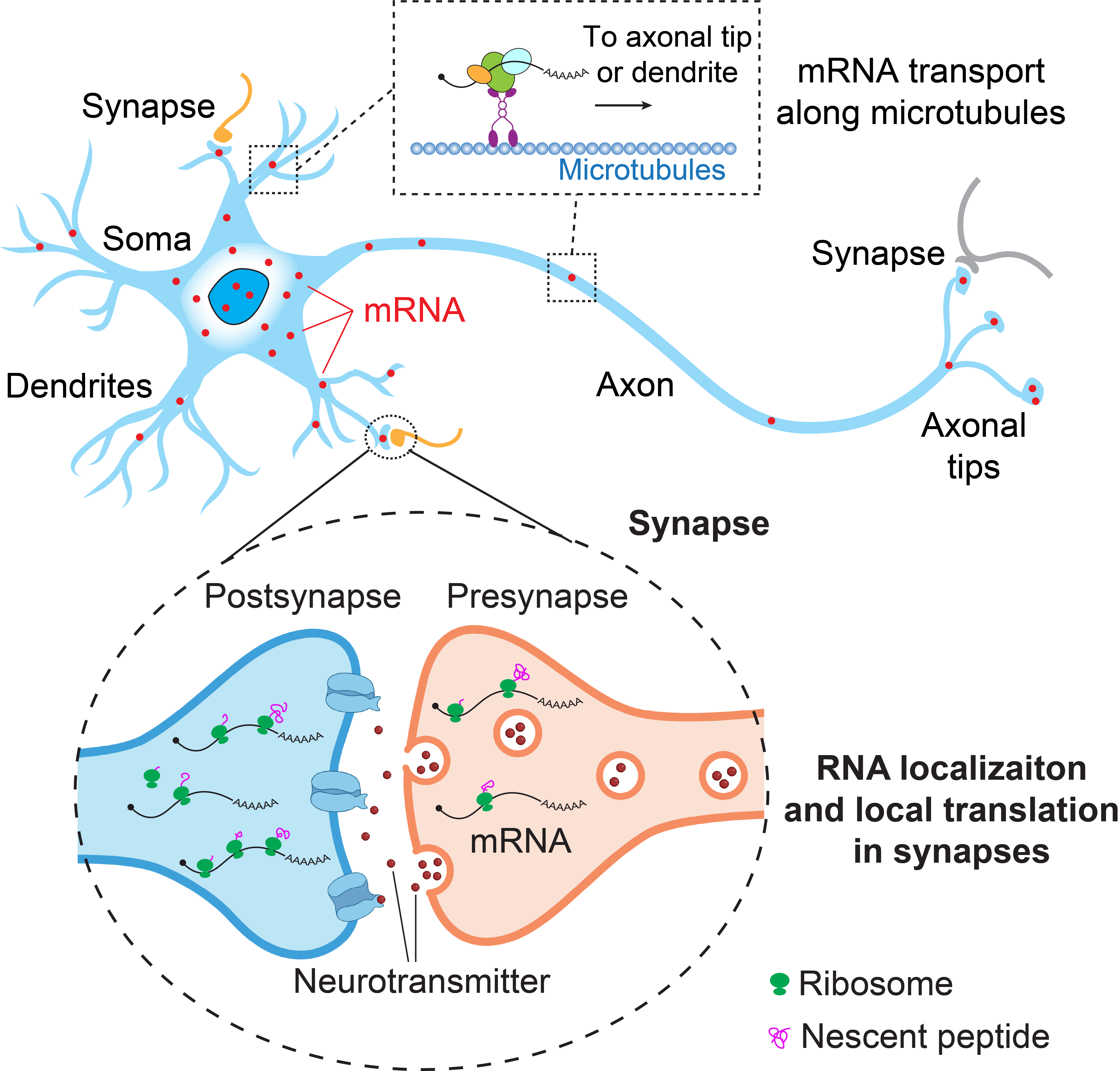
Spatial RNA localization is a key epigenetic mechanism regulating the spatiotemporal gene expression. This mechanism is particularly important in neurons and plays a crucial role in neuronal functions including neurite regeneration and synaptic plasticity which is the cellular basis underlying learning and memory and neurite circuit formation. Aberrant RNA localization has been observed in neurological disorders including Amyotrophic Lateral Sclerosis (ALS), Frontotemporal Dementia (FTD), and Spinal Muscular Atrophy (SMA) neurodegenerative diseases. However, the regulatory mechanisms, functional roles, and pathological relevance of spatial RNA localization in neurons remain poorly understood, largely due to limited tools. The Han lab aims to address these knowledge caps by applying and developing cutting-edge synthetic biology, chemical biology, and CRISPR technologies to profile and manipulate the spatial transcriptome. Our long-term goal is to develop novel therapeutic strategies to treat neurological disorders including neurodegenerative diseases and substance use disorders via spatial transcriptome engineering.
Current Research Directions
Direction 1: Engineer better programmable RNA-targeting systems
Our lab currently uses Class 2 type VI CRISPR-Cas13 systems for manipulating spatial RNA localization to study its biological functions. CRISPR-Cas13 is an RNA-guided, RNA-targeting system which uses guide RNA (gRNA) to direct the ribonuclease Cas13 to cleave the target RNA. Mutating the ribonuclease domain of Cas13 creates a catalytically inactive (dead) Cas13 variant (dCas13), allowing targeted RNA binding without RNA cleavage. However, dCas13 is relatively large for in vivo applications and often exhibits suboptimal RNA-targeting efficiency. We are actively doing protein engineering to develop new programmable RNA-targeting systems that are more compact with robust RNA-targeting efficiency for studying RNA biology.
Direction 2: Comparative spatial transcriptome profiling
Spatial RNA localization has been found to play a crucial role in numerous essential neuronal functions and processes, including neurite regeneration, synaptic plasticity, neural circuit formation, and learning and memory. Our lab is interested in establishing novel platforms for comparative spatial transcriptome profiling to identify key RNA localization events participating in these neuronal functions and processes. We will collaborate with other labs at IU to apply the platforms for comparative spatial transcriptome profiling in multiple biological systems, including: 1) microfluidic device-based axonal injury model of neurite regeneration, 2) Drosophila melanogaster models of learning and memory and neurodevelopmental disorders, and 3) mouse models of opioid use disorder.
Direction 3: Functional study of the spatial transcriptome
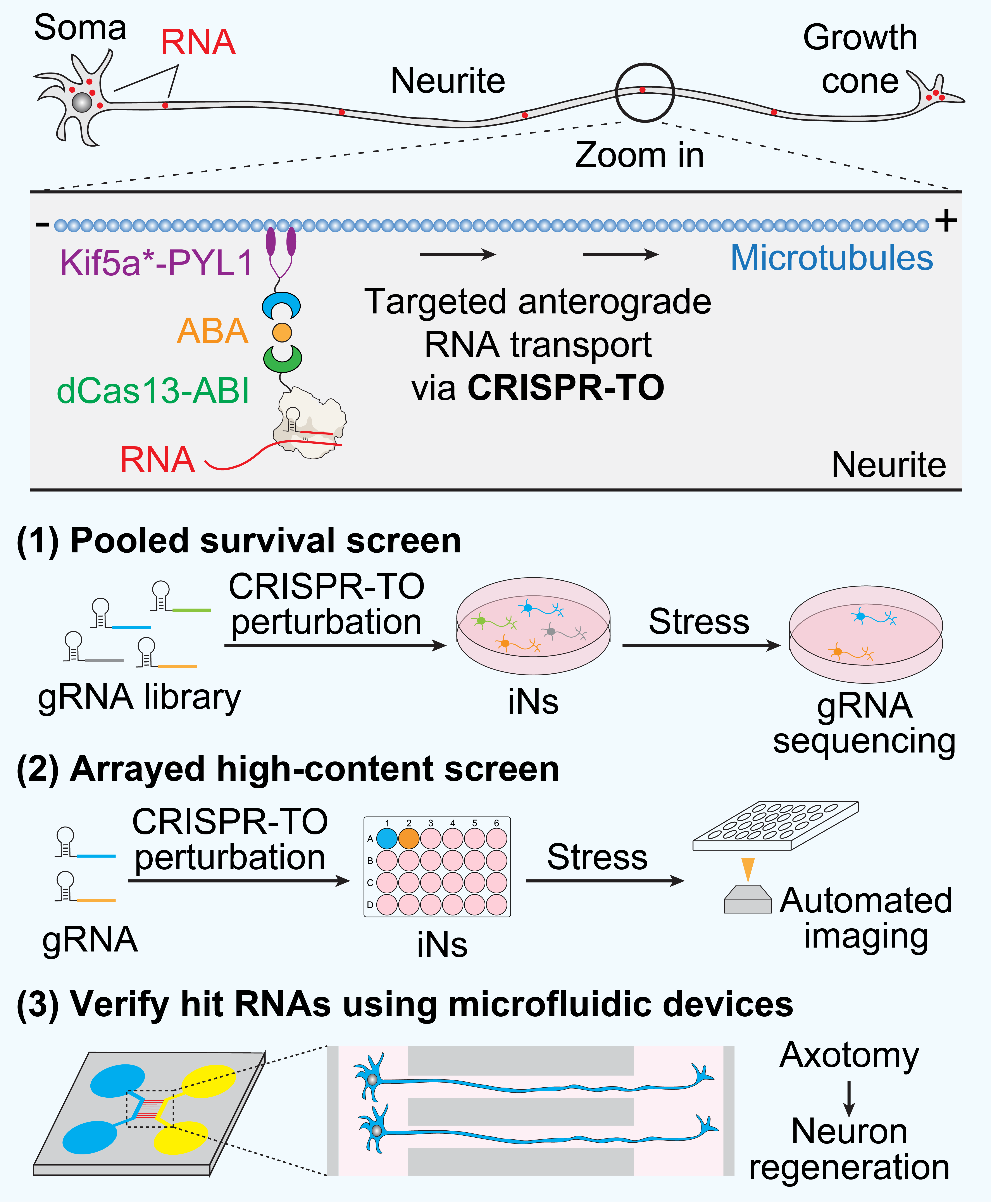
Functional study of the spatial transcriptome remains in its infancy, largely due to a lack of tools for perturbing endogenous RNA localization in living cells and neurons. Traditional methods for manipulating RNA localization require genome engineering to remove regulatory elements or insert RNA aptamers for tethering to specific subcellular locations, which are low-throughput and laborious to be applied in neurons. To overcome these limitation, Dr. Han developed the CRISPR-mediated transcriptome organization (CRISPR-TO) technology, which enables the programmable control of the spatial transcriptome in live cells and neurons. The Han lab is actively optimizing the CRISPR-TO technology to improve its performance for high-throughput screening and in vivo applications. Our lab will apply the CRISPR-TO technology for high-throughput screening to investigate the functions of the spatial transcriptome, particularly focusing on key spatial RNA localization events identified in essential neuronal functions and processes listed in Direction 2.
